#!/usr/bin/env python2.7
# initialize PETSc
import sys, petsc4py
petsc4py.init(sys.argv)
from petsc4py import PETSc
import numpy as np
# import pyCTQW as qw
import pyCTQW.MPI as qw
# enable command line arguments -t and -N
OptDB = PETSc.Options()
N = OptDB.getInt('N', 10)
t = OptDB.getReal('t', 20)
# get the MPI rank
rank = PETSc.Comm.Get_rank(PETSc.COMM_WORLD)
if rank == 0:
print '1P 3-cayley Tree CTQW\n'
# initialise a 10 node graph CTQW
walk = qw.Graph(N)
# Create a Hamiltonian from a file.
# A defect of amplitude 3 has been added to node 0
walk.createH('../graphs/cayley/3-cayley.txt','txt',d=[0],amp=[3.])
# create the initial state (1/sqrt(2)) (|0>+i|1>)
init_state = [[0,1.0/np.sqrt(2.0)], [1,1.0j/np.sqrt(2.0)]]
walk.createInitState(init_state)
# set the eigensolver properties. Note that
# Emin is exact, whereas and Emax has been over-estimated.
walk.EigSolver.setEigSolver(tol=1.e-2,verbose=False,emin_estimate=0.,emax_estimate=10.)
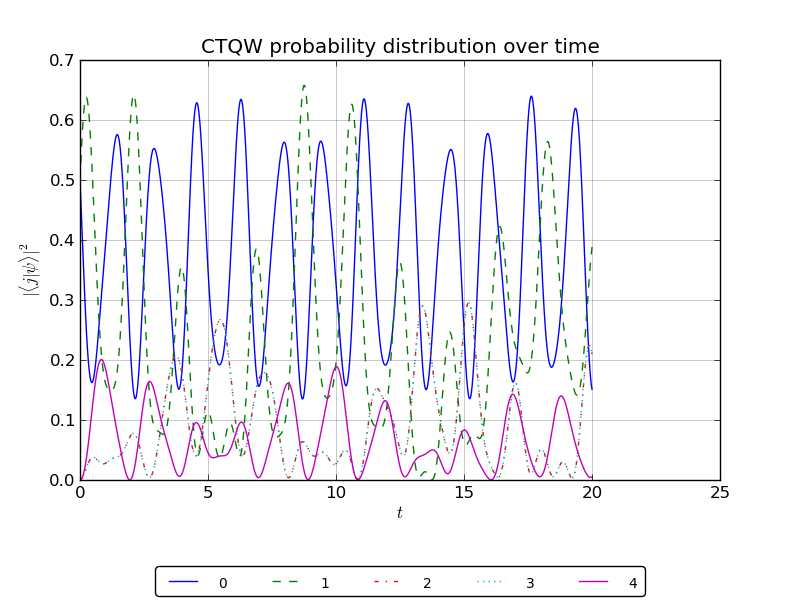
# create a handle to watch the probability at nodes 0-4:
walk.watch([0,1,2,3,4])
# Propagate the CTQW using the Chebyshev method
# for t=20s in timesteps of dt=0.01
for dt in np.arange(0.01,t+0.01,0.01):
walk.propagate(dt,method='chebyshev')
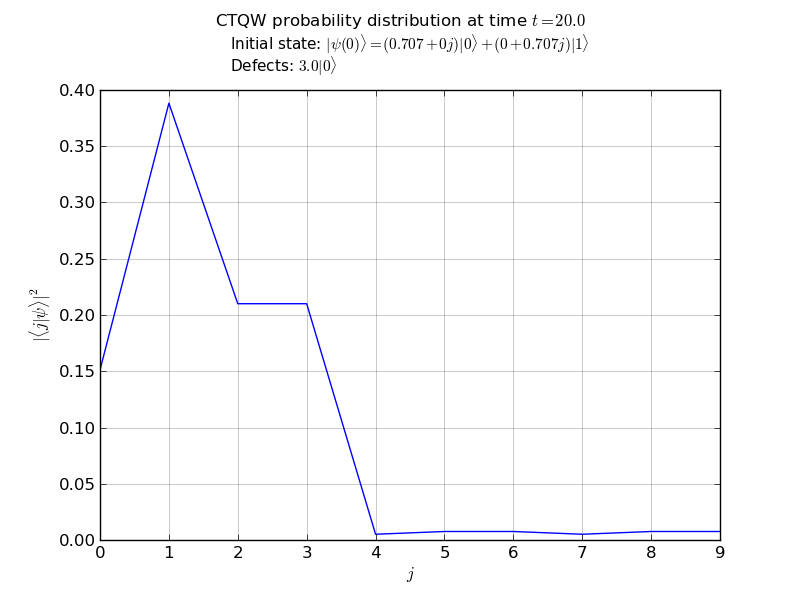
# plot the marginal probabilities
# after propagation over all nodes
walk.plot('out/1p_3cayley_plot.png')
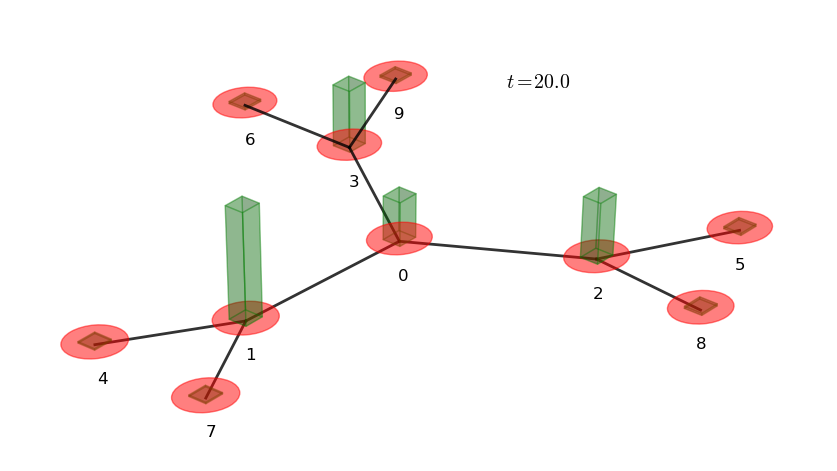
# plot a 3D graph representation with bars
# representing the marginal probabilities
walk.plotGraph(output='out/1p_3cayley_graph.png')
# plot the probability over time for the watched nodes
walk.plotNodes('out/1p_3cayley_nodes.png')
# destroy the quantum walk
walk.destroy()